More SynthNet goodness today! First off, I finished up the changes to the code that ensure all parts of SynthNet were relatively in sync with each other. With one of my next big tasks being focusing on rate and temporal coding, timing within the system needs to be correct to support exact oscillating frequencies of action potentials, as well as resonance. There were some significant changes to the code, so I needed to retest most parts of the entire program again. That took up pretty much the month of August and beginning of Sept – all works well though!
Mutative Madness!
Before I took the next step and jumped into the neural coding work, I wanted to program the functionality that allows for the mutation of SynthNet’s virtual DNA, accommodating evolution experiments. I finished up the mutation engine itself a couple nights ago, and am starting on the interface portion that will allow external programs to perform artificial selection experiments by monitoring the effectiveness of a DNA segment – either continuing its mutation if successful, or discarding the genetic line and returning to a previous if less successful.
Below can be seen examples of the effects of mutation performed on a virtual DNA segment. The first picture shows a network grown with the original, manually created DNA (the segment used in my classical conditioning experiment)
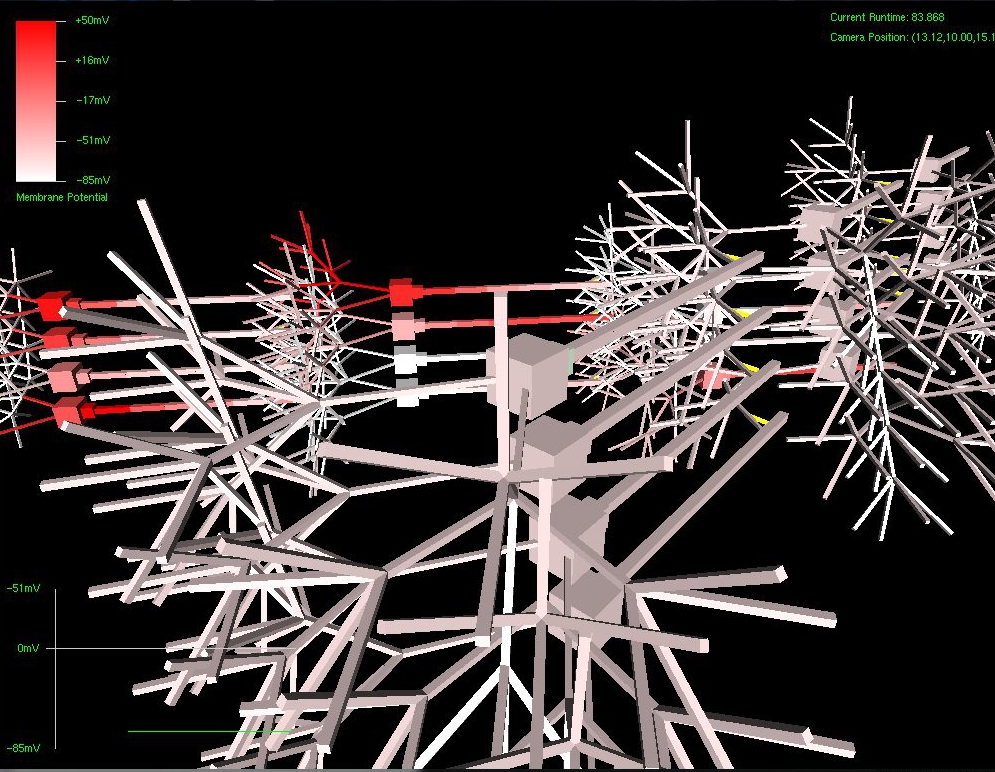
The next set of pictures show the results of a neural network grown using DNA that has undergone a .5% – 2% amount of mutation. Most were beautiful to look at, but the final two pictures were also completely functional, supporting the proper propagation of action potentials and integration of synaptic transmission – only with an entirely novel configuration!
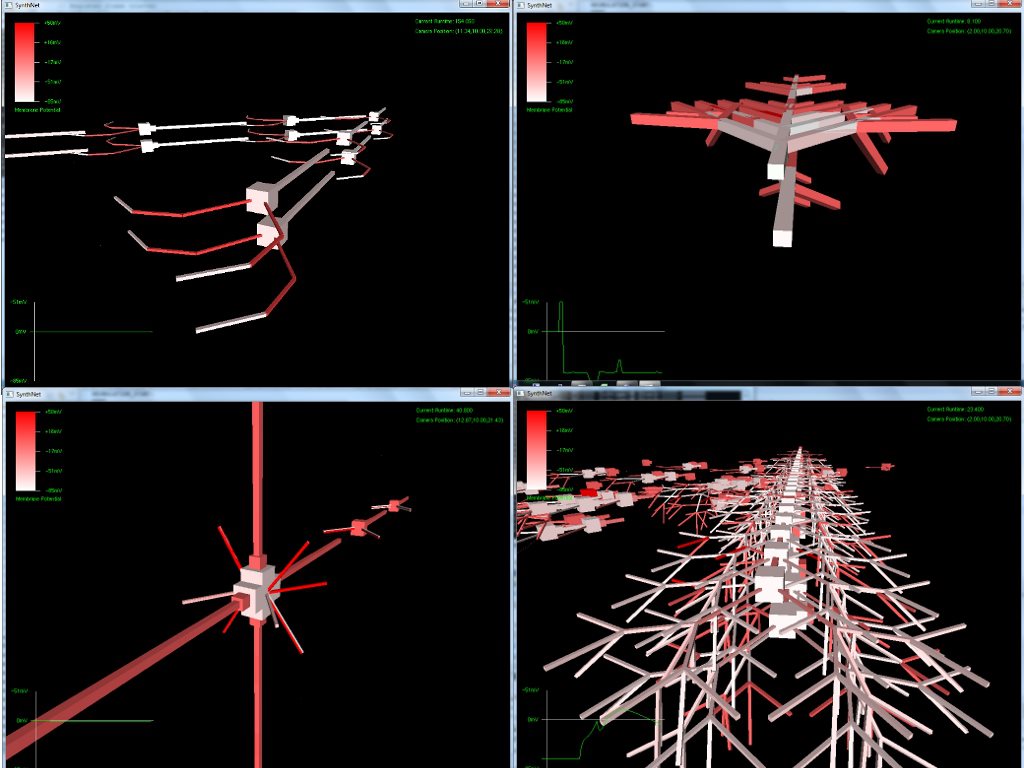
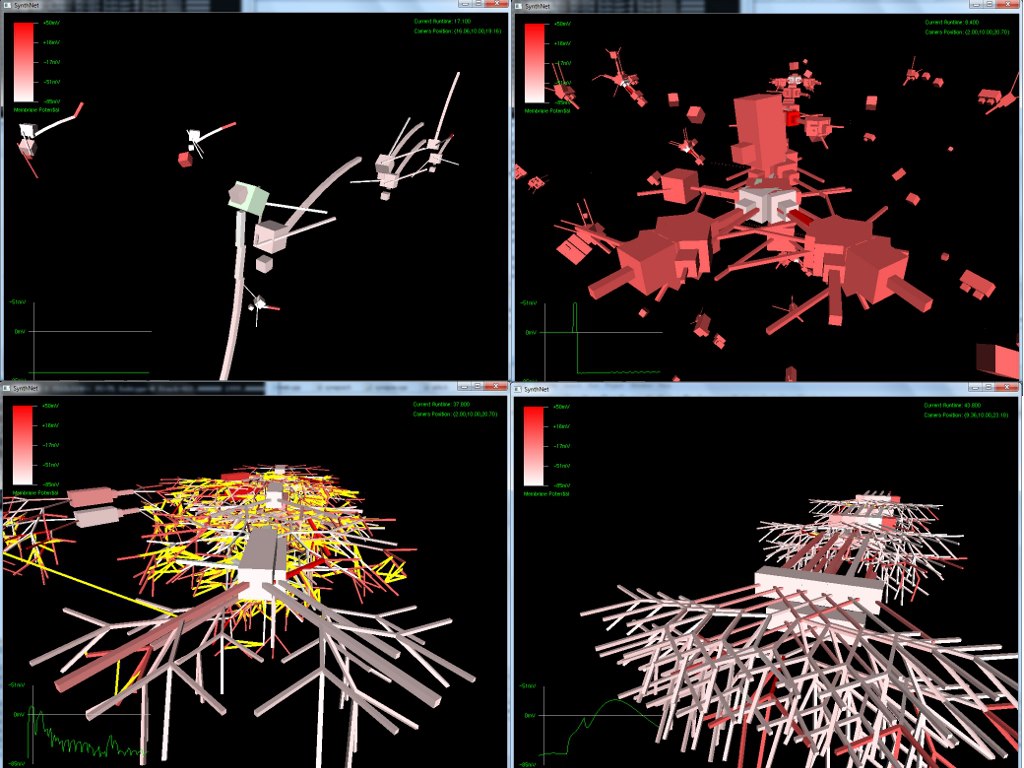
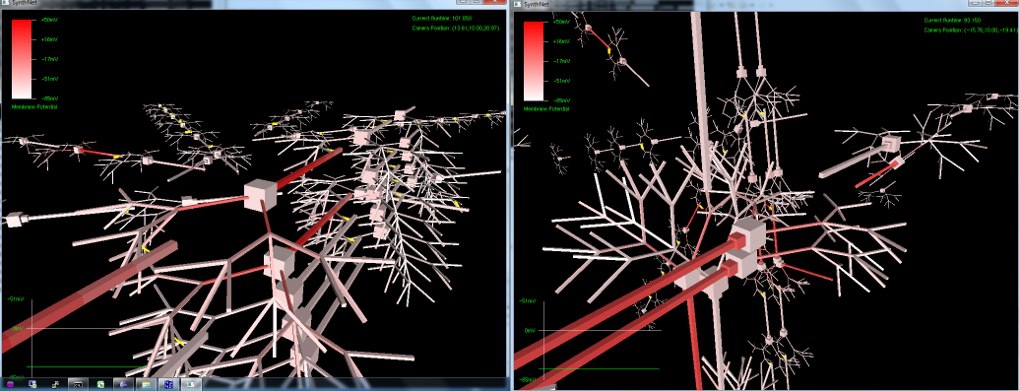
Really amazing to look at (I think!) Currently, SynthNet DNA can be exposed to the following types of errors in its genomic sequences:
- Deletion – Segments are removed entirely
- Duplication – Segments are copied in a contiguous block
- Inversion – Segments are written in reverse
- Insertion – Segments are moved and inserted into a remote section
- Translocation – Akin to Insertion, but two segments are swapped with each other
- Point Mutations – Specific virtual nucleotides are changed from one type into another
Currently, these operations result in in-frame mutations. It was actually easier to allow frameshifts to occur – however, SynthNet DNA is more sensitive to framing, since whereas a biological read frame is across a codon (3 nucleotides), SynthNet DNA is variable from 1-6 virtual nucleotides. When I allowed frameshifting, the results were high in nonsense mutations, which prevented almost any meaningful growth of neural structures.
Very excited with how things are turning out. If, as seen in the last two pictures, we can get such novel pathway growth with a simple random mutation, I can’t wait to start the artificial selection routines and watch the results unfold!
 Hello - and thanks for visiting my site! I maintain ToniWestbrook.com to share information and projects with others with a passion for applying computer science in creative ways. Let's make the world a better and more beautiful place through computing! | More about Toni »
Hello - and thanks for visiting my site! I maintain ToniWestbrook.com to share information and projects with others with a passion for applying computer science in creative ways. Let's make the world a better and more beautiful place through computing! | More about Toni » 







Leave a Reply