SynthNet
 Click Here For Latest News / Dev Blog
Click Here For Latest News / Dev Blog
SynthNet is a (very) on-going artificial intelligence research project I started back a number of years ago as the more simple TFNN project – then restarted in its current incarnation in 2011. The ultimate goal is a true, functional model of the biological neural network in software grown using virtual, evolved DNA. While this is an incredibly lofty goal, the project serves as more of a learning opportunity for me (and anyone else interested). I am not a neuroscientist or a genetics expert by any means, but I am very fascinated by both topics, especially surrounding integrating principles of neurophysiology into a virtual environment, for academic, biomedical, and unforeseen technological singularity opportunities.
Watch a demonstration of SynthNet in action here!
Currently, SynthNet is Capable of the Following:
- Compiling SynthNet genetic code into virtual DNA, which can be used to direct neuronal growth, network configuration, protein synthesis, receptor growth, structure differentiation, and anything else SynthNet is capable of. An example of this can be seen both with growth and CamKII functionality in the phase 1 demonstration video above. Each neural structure runs a Genetic Virtual Machine.
- Processing physical attributes about neural structures, such as positioning and morphology, intracellular resistance, plasma membrane capacitance and resistance, etc. This functionality also allows for the emulation of myelination and complex signal integration.
- Processing substance concentrations and charges, both in the intra and extracellular spaces. These substances include ions, neurotransmitters and modulators, as well as proteins.
- Processing channels embedded in the plasma membrane. Functionality, including permeability calculations, of these channels are dictated by virtual “protein subunits” which control ion permeability type and amount, gating (voltage/ligand/activation/inactivation), ligand agonists/antagonists, etc. Calculations make use of standard equations such as Hodgkin-Huxley.
- Processing synaptic connections between pre and post-synaptic neurons, as well as neurotransmitter vesicle release. Vesicle fusion probabilities are dictated by a number of means, including poisson and binomial estimation via calcium concentrations in microdomains as well as structure wide.
- Calculation of membrane and resting potentials via GHK, and spread of electrotonic potential across neural structures via cable method.
- Genetic Mutation Engine, capable of performing common mutations at a specified probability level. Mutation types include: point mutations, deletions, duplications, inversions, insertions, and translocations.
- TCP/IP Peripheral Nervous System, capable of allowing sensory input/output to any network connected device (robots, cameras, microphones, who-knows-what), as well as (soon to be) distributed sections of the neural network across multiple machines. Additionally allows access to the genetics and genetic mutation engine, allowing for the upload/download of DNA, control over mutation, and (soon to be) recombination.
- Virtual parallel processing, multithreading, and substance sharing across structures.
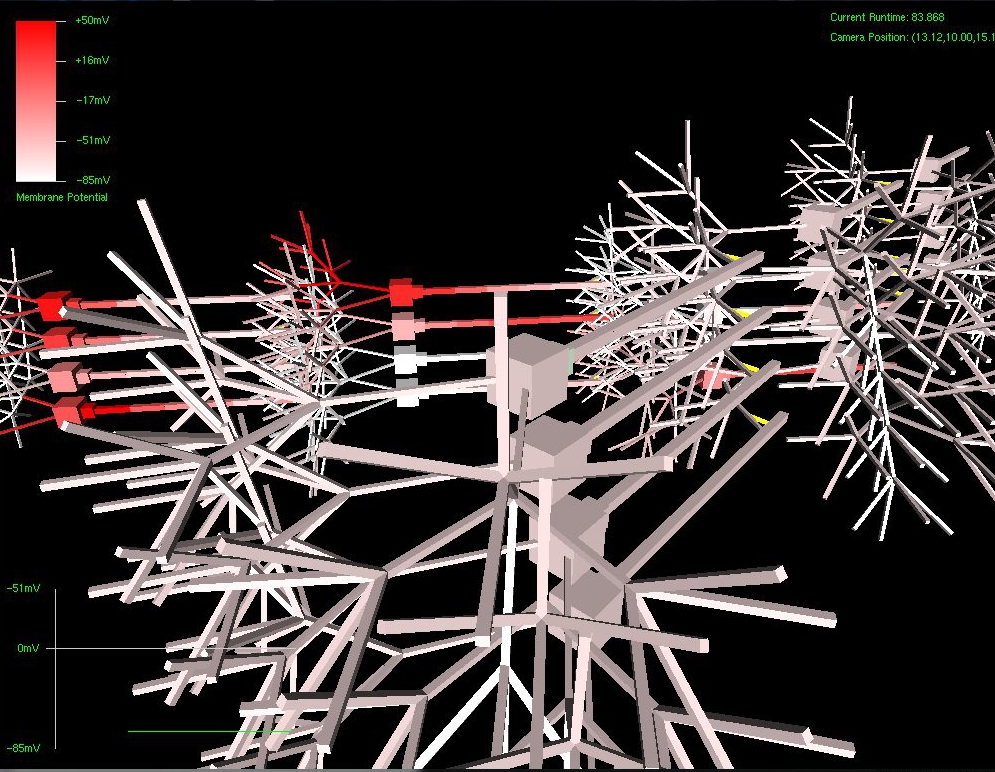
- Real time, “virtual fMRI” visualization engine that renders a physical view of the virtual SynthNet network. Membrane potentials are displayed as color gradients, and synaptic clefts by yellow lines. Additionally, a graph representation showing potential history of a specified neural structure is also displayed for detailed study of electrical activity.
- Lots more!
Still To Do:
- Create experimentation client (in Python) to connect to a SynthNet via the TCP/IP PNS engine, and run specified experiements, such as artificial selection to demonstrate evolution capabilities – results recorded via database connection.
- Add recombination functionality to the genetics code, aligned to frames and entry points, which will allow the integration of two DNA segments, including via the TCP/IP engine from a remote source to simulate sexual reproduction
- Write genetics to emulate different types of neural coding, including rate/time/spike/etc.
- Experiment with manually creating more complex genetic code, taking advantage of neural coding work, to emulate the storage and retrieval of declarative memories.
- A number of other projects – to be listed soon!
Thanks for checking on the project, please write with any questions or comments!
 Hello - and thanks for visiting my site! I maintain ToniWestbrook.com to share information and projects with others with a passion for applying computer science in creative ways. Let's make the world a better and more beautiful place through computing! | More about Toni »
Hello - and thanks for visiting my site! I maintain ToniWestbrook.com to share information and projects with others with a passion for applying computer science in creative ways. Let's make the world a better and more beautiful place through computing! | More about Toni » 



