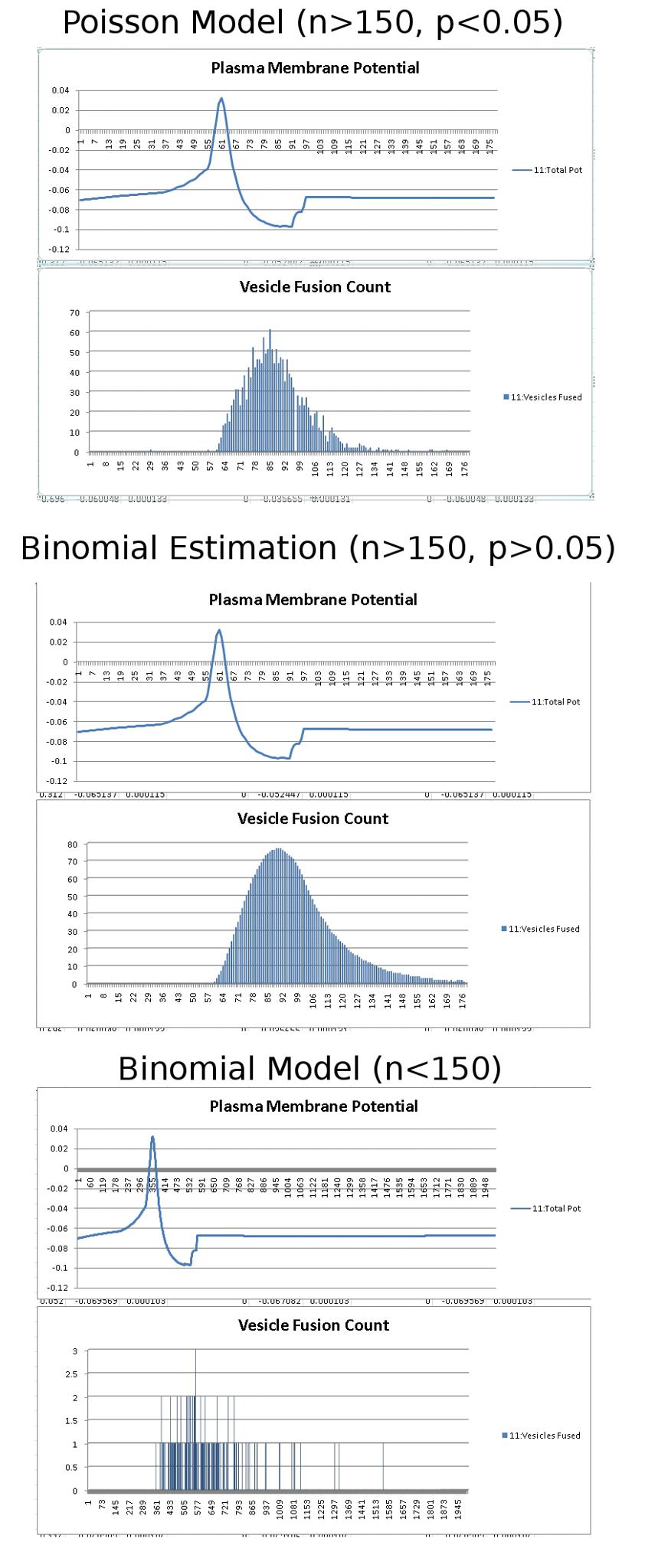
After finishing up phase 1 of SynthNet, I came to the conclusion that I really missed updating the blog. When I get involved in a project, I tend to get wrapped up in it (more accurately – completely and ridiculously obsessed where it takes over my life) and other things drop off the radar. However, I’ve decided I want to make a real effort to not get AS wrapped up in projects, and remember to give the blog some love.
New and Improved!
As I went to write my first article after recording the SynthNet video, I also noticed the blog was looking a little tired. They’d also made a number of improvements in WordPress since when I first installed everything, so I decided to take the leap, get a shiny new template, and put some new life into it. I think it’s definitely an improvement – hope you enjoy it!