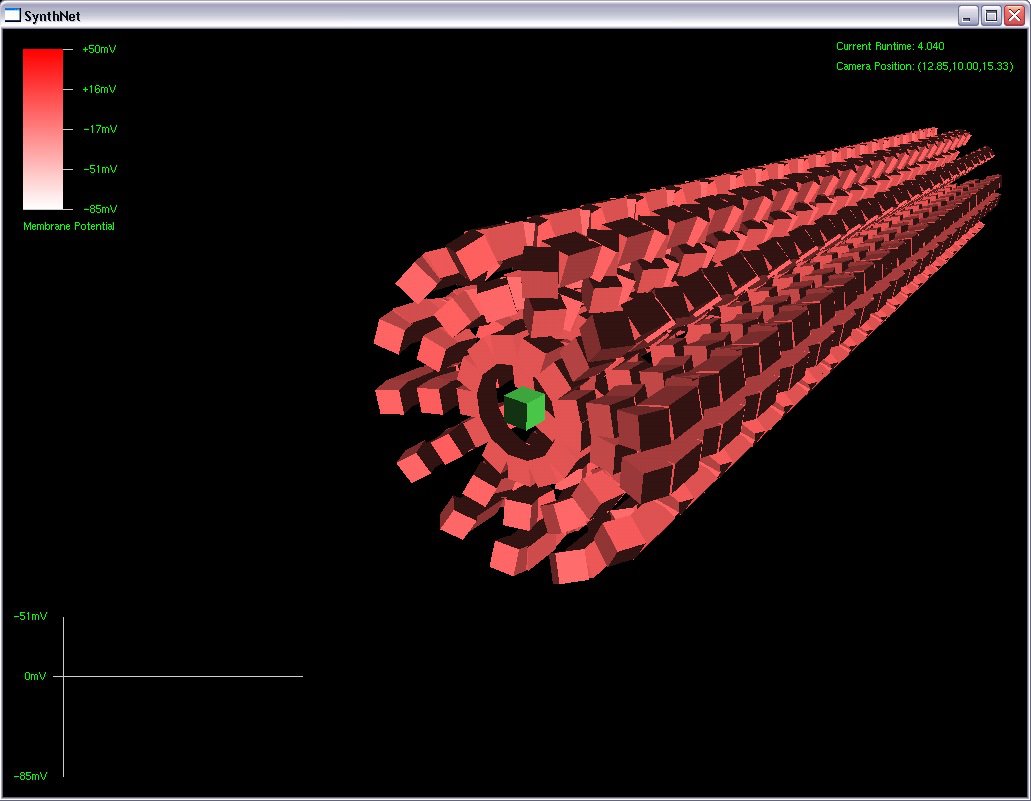
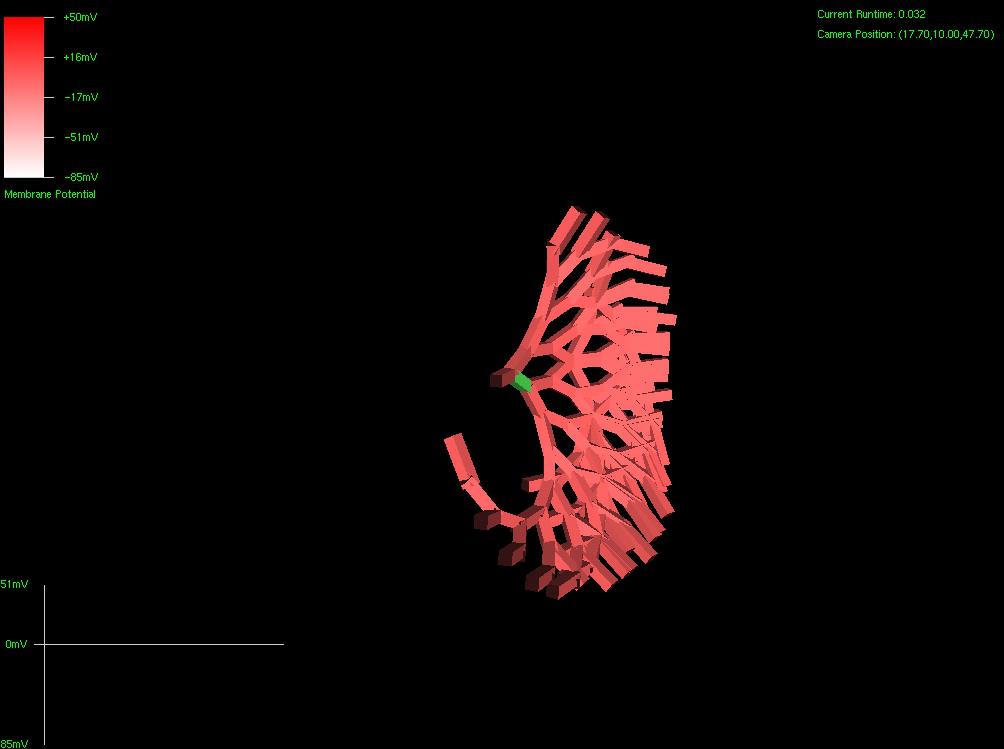
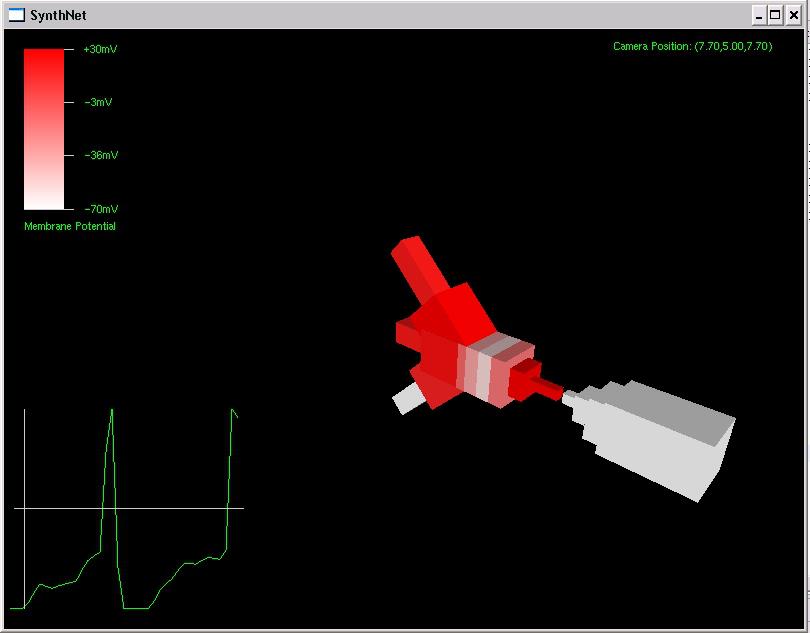
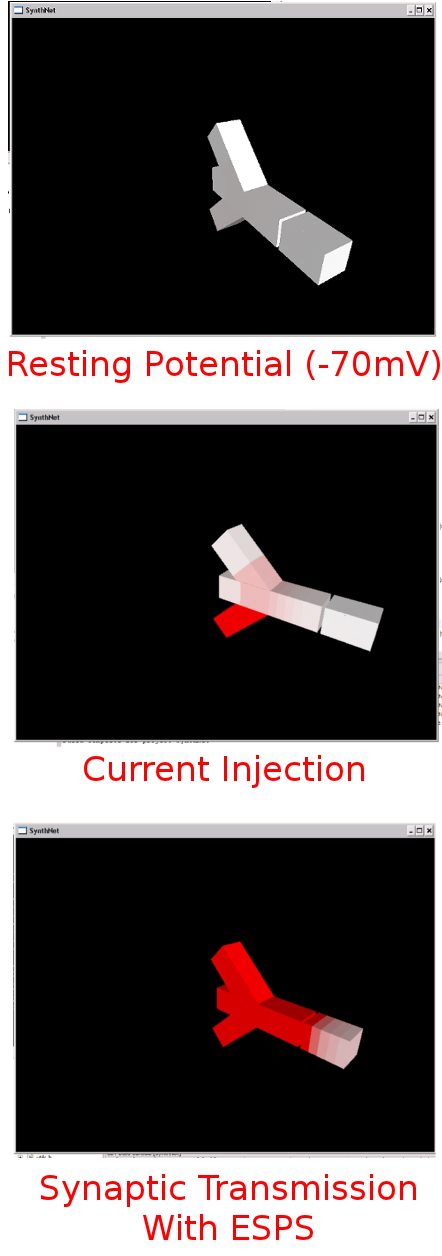
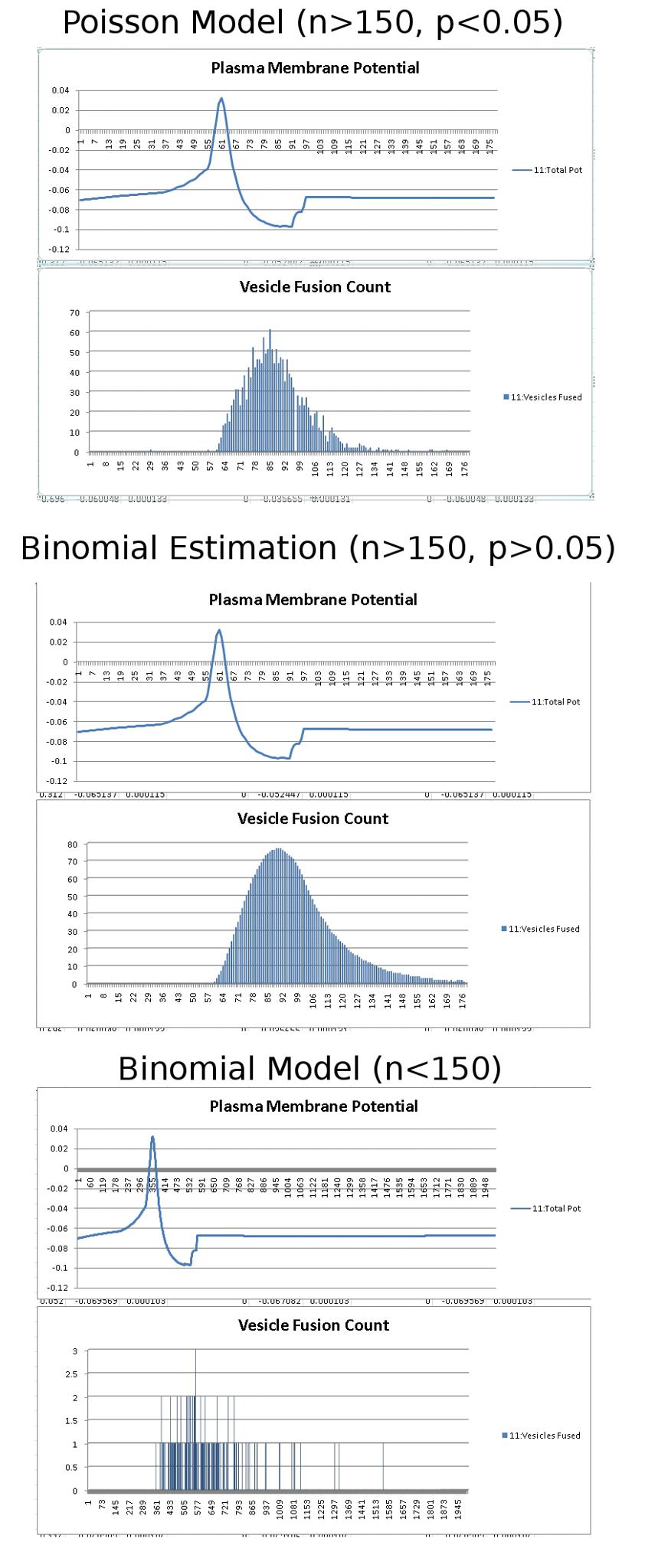
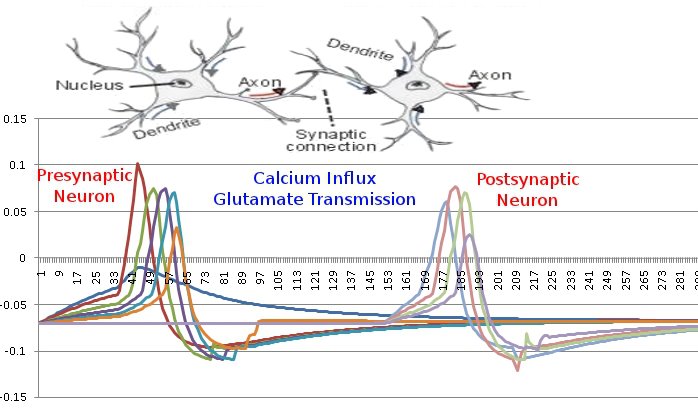
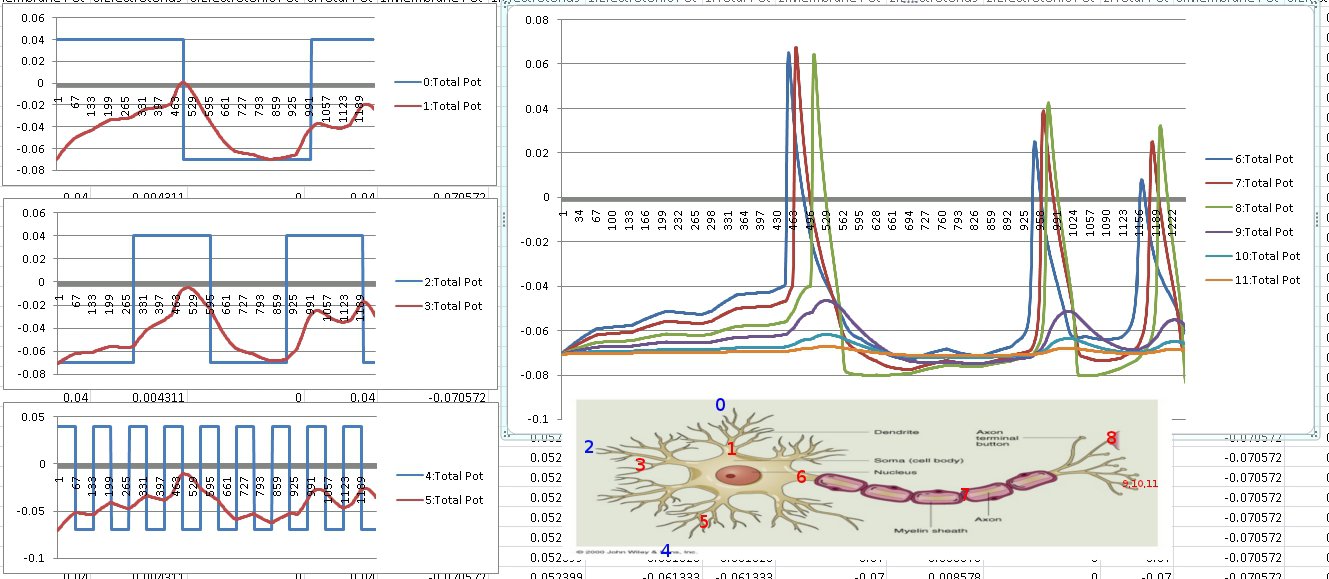
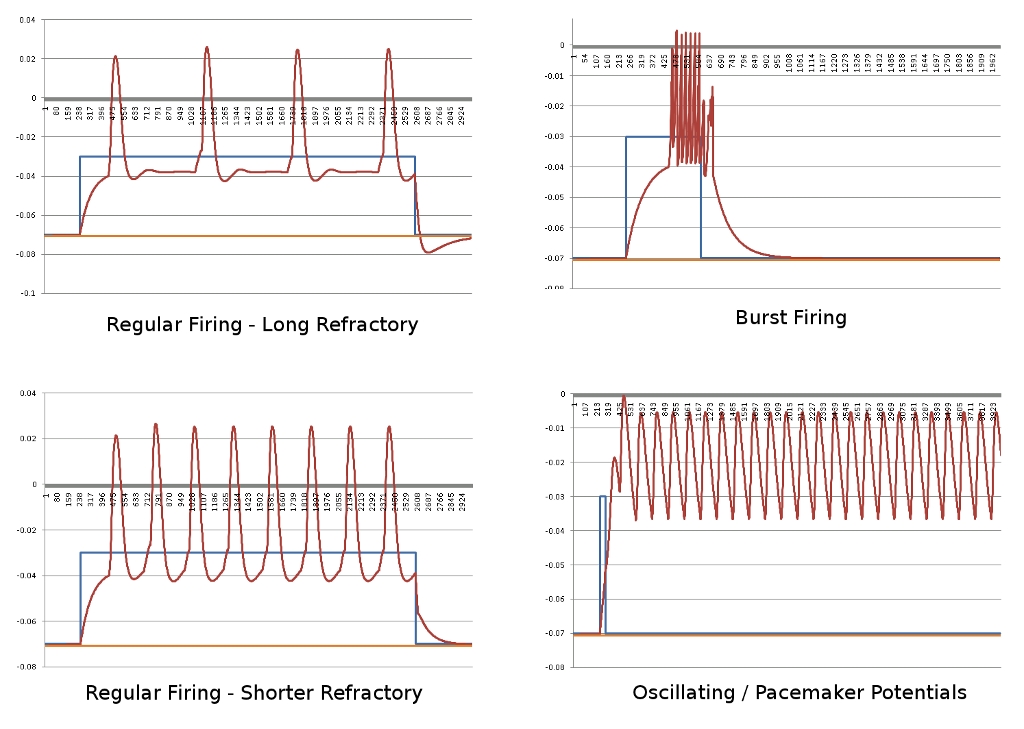
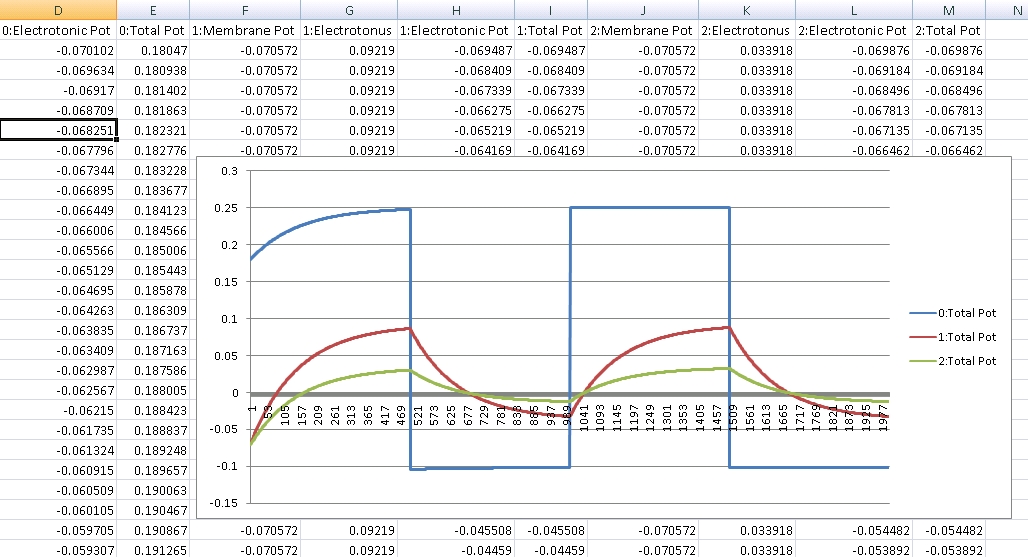
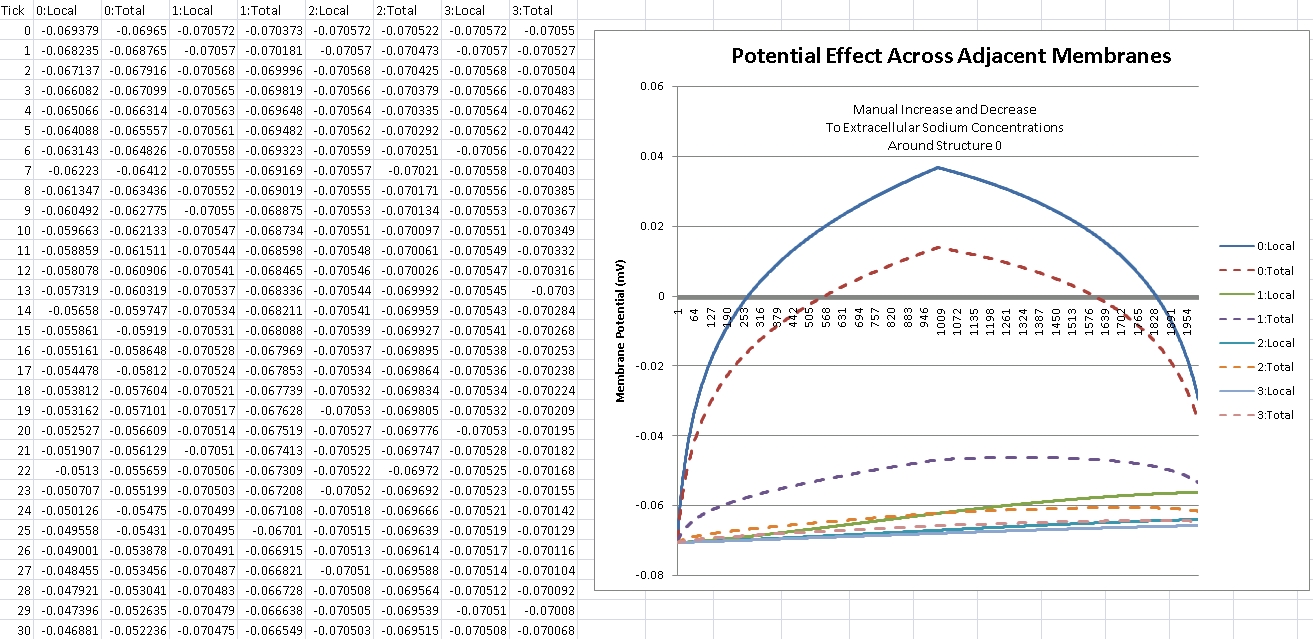
After about 8 hours of programming, I got mitosis and stem cell differentiation working in the genetic engine yesterday. Pictured is a 1840 neuron columnar nucleus, grown in a linear fashion initially, then radiating outward via differentiating mitosis. SynthNet is now able to regulate growth (so it isn’t cancerous), as well as direct growth based on protein signaling markers, allowing it to grow differentiated structures.
-
About
-
 Hello - and thanks for visiting my site! I maintain ToniWestbrook.com to share information and projects with others with a passion for applying computer science in creative ways. Let's make the world a better and more beautiful place through computing! | More about Toni »
Hello - and thanks for visiting my site! I maintain ToniWestbrook.com to share information and projects with others with a passion for applying computer science in creative ways. Let's make the world a better and more beautiful place through computing! | More about Toni »
-
My Highlighted Projects
- Cloudburst Connection - our newest game!
- Shredz64 - Guitar rhythm game for C64
- Whirlwind - NES compatible FPGA Core
- PALADIN - Metagenomic characterization
- RepeatFS - File system for scientific reproducibility
- SynthNet - Neural network simulation
- NetPaint - Text-based drawing
- GitHub - View all projects
Recent Comments
- Toni on Transferring files to the Osborne 1
- CloudInMyHead on Transferring files to the Osborne 1
- Kelsey Travis on Converting Your Corporate Intranet to Drupal
- Lyle on Transferring files to the Osborne 1
- Ander on Evolution Experimentation Module Complete
Archives